Last Updated on
Sentinel surveillance of imported dengue in returning travelers: trends and virus evolution
Background
Dengue, a mosquito-borne viral infection, has evolved into a major international public health concern putting worldwide more than 2.5 billion people at risk for infection.
Over the last decades the virus has spread around the globe leading to increased frequency and magnitude of dengue epidemics. Factors for these developments are complex and result from virus evolution (towards greater epidemic potential), ecological and climate changes (affecting vector biology), and population dynamics (urbanization and increased mobility) of the natural human host.
Dengue is an important cause of fever in returning travelers, and the frequency of importation of dengue into currently non-endemic areas via travelers is increasing.1 This importation of the virus to non-endemic areas is accompanied by the introduction or already presence and rapid expansion of potential mosquito vectors. The potential threat of this development has been highlighted by the recent cases of autochthonous dengue transmission in North America and Europe.
Surveillance is the single most important tool for identifying the geographical expansion and dynamic of dengue as well as the changing molecular epidemiology of the virus worldwide. Unfortunately, global surveillance for dengue remains fragmentary and current surveillance systems and control efforts have proven insufficient to combat dengue in endemic countries and to prevent the introduction into previously uninfected regions (including Europe).
To fill the currently existing scientific gaps in dengue epidemiology the international research initiative “![]() DengueTools” has been launched. Funded by the “European commission’s Seventh Framework Programme (FP7)” the initiative will focus on dengue surveillance, prevention, and the risk of introduction to uninfected areas. “TropNet”, the “European Network for Tropical Medicine and Travel Health” has agreed to collaborate with the DengueTools initiative by participating in the FP7 dengue research project. The research focus of TropNet will be on European travelers, who get infected abroad and import dengue infections to Europe:
DengueTools” has been launched. Funded by the “European commission’s Seventh Framework Programme (FP7)” the initiative will focus on dengue surveillance, prevention, and the risk of introduction to uninfected areas. “TropNet”, the “European Network for Tropical Medicine and Travel Health” has agreed to collaborate with the DengueTools initiative by participating in the FP7 dengue research project. The research focus of TropNet will be on European travelers, who get infected abroad and import dengue infections to Europe:
The use of travelers as sentinel to enhance surveillance is an effective public health tool that has proven useful in the monitoring of many diseases. Travelers might well be seen as early indicators of outbreaks that could go otherwise unnoticed at certain travel destination countries. A well-characterized viral isolate from a traveler could presage the appearance of a new serotype or strain in a geographic area, where respective local institutions are not existing or not capable of performing sophisticated laboratory investigations. Additionally complete genomic sequencing of dengue virus strains isolated from travelers to different geographical locations over time will offer the opportunity to understand the genetic stasis, possible selection pressure, and evolution in the dengue genome.
Significance of this study
1. Contribution to the global epidemiological picture of dengue
Surveillance is the single most important tool for identifying the geographical expansion of dengue, variations over time, and the changing epidemiology of dengue worldwide. Unfortunately, global surveillance for dengue remains fragmentary. Earlier recognition of dengue could be enhanced by targeting certain vulnerable populations (sentinel) for surveillance. The use of sentinel groups to enhance surveillance is an effective public health tool that has proven useful in the monitoring of many diseases. International travelers can potentially serve as such a sentinel group for dengue. Travelers might well be seen as canaries (‘the sentinel birds in the coal mines’), as early indicators of outbreaks that could go otherwise unnoticed at certain travel destination countries: Travelers transport the dengue viruses from geographic areas in (sub-)tropical developing countries, where limited laboratory facilities exist, to countries of the North with laboratories that can identify viruses. Detection of even single patients with a disease from a geographically unexpected area or of small numbers of patients with similar unusual events might have immediate significance. Such sentinel surveillance of travelers would augment, not replace, existing national public health surveillance systems. Surveillance of travelers can provide an additional layer of international surveillance effort. An added benefit is that a well characterized viral isolate from a traveler could presage the appearance of a new serotype or strain in a geographic area. Because most travelers from temperate areas are immunologically naïve (will not have had prior experience with dengue viruses) and can provide details of their travel itinerary, it may sometimes be easier to determine the most probable time and place of exposure in travelers than in local residents.
Molecular epidemiological studies are crucial in determining the transmission patterns of dengue viruses and tracking the spread of dengue around the world. As travelers return from any of the countries where dengue is currently endemic, they potentially reflect the evolving epidemiology (strain distribution and circulation) of dengue. Complete genomic sequencing of dengue virus strains collected from different geographical locations and isolation years via returning travelers offers the opportunity to understand phylogenetic trees, possible selection pressure sites, and evolution in the dengue genome that may pose an increased risk for epidemic potential. Little is known about the genetic differences of full-length viral genomes of dengue viruses. Complete genomic sequencing of dengue virus strains collected from different geographical locations and isolation years offers the opportunity to understand the genetic stasis, possible selection pressure sites and evolution in the dengue genome. The Genome Institute in Singapore has developed a chip-based approach to viral sequencing, to increase efficiency and enable large numbers of genomes to be completed, together with a web-based interpretation tool. Our results can be compared and shared with “DengueInfo”. ![]() DengueInfo is a web portal and database on dengue research integrating the growing number of complete genome sequences of dengue virus with relevant literature and curated epidemiological information. Our data would be an additional resource for the dengue community, providing standardized and high quality information about dengue phylogenetic studies and genomics from locations all around the world.
DengueInfo is a web portal and database on dengue research integrating the growing number of complete genome sequences of dengue virus with relevant literature and curated epidemiological information. Our data would be an additional resource for the dengue community, providing standardized and high quality information about dengue phylogenetic studies and genomics from locations all around the world.
2. Surveillance of dengue importation to non-endemic regions
Viremic humans, whether troops, migrant workers, tourists, business travelers, refugees, or others, can carry the virus into new geographical areas, which might lead to outbreaks, if a competent vector inhabits the new area. Although a great deal of effort is taken to prevent the spread of dengue viruses in infected mosquitoes by implementing mosquito abatement programs at international airports and spraying adulticides in passenger cabins of arriving aircraft, mosquitoes as agents of spread are probably overrated and viremic travelers are the most likely source of the importation of dengue viruses.
Therefore when assessing the risk of introducing dengue to non-endemic regions (like Europe) the key focus of surveillance is – next to the evaluation of the local prevalence and distribution of potential mosquito vector species – the evaluation of the extent of imported dengue via travelers.
3. Evaluation of clinical dengue manifestations in travelers
The evaluation of the clinical manifestations of dengue in travelers is of particular interest as travelers from non-endemic regions represent an immunological naïve population which might exhibit differences in clinical presentations when compared to populations from dengue endemic regions. There is a debate on whether severe haemorrhagic dengue manifestations are due to repeated contact with (different) serotypes, caused by sequential dengue infections. The (i) proposed antibody-dependent enhancement (ADE) theory, demonstrated in in-vitro experiments, which assumes that primary infections give rise to non-neutralizing antibodies which may enhance the entry of virus into Fc-γ receptor-positive cells, or (ii) the ‘aberrant T-cell response’-theory were questioned as sole explanantions because dengue haemorrahgic fevers were also observed among primary cases. Virulence of genotypes and complex interaction of host and other factors may influence the clinical outcome of dengue infection. Hence, a continuum from non-clinical to fatal cases has been proposed, leading to a revised classification of severe dengue manifestations to facilitate appropriate triage and clinical management. The latest WHO dengue classification from 2009 thus contains a pragmatic approach of defining dengue disease for practical triage and clinical management. In this study, the new classification will be comparatively assessed with the old one from 1997 for returning travelers, according to the case classification by severity, i.e. ‘Dengue without warning signs’, ‘Dengue with warning signs’ and ‘Severe dengue’. If data quality and number of patients allows, a comparison of the clinical classification will be compared with dengue cases in Sri Lanka among an endemic population of patients.
In addition to the clinical data of imported dengue cases, complete genomic sequencing of the respective virus strains will be conducted. This might allow the correlation of specific clinical manifestations to specific differences within the viral genome.
Objectives
- to describe the epidemiological trends of dengue infections imported to Europe by returning travelers
- to describe the full clinical spectrum of dengue disease imported to Europe by returning travelers
- to report the phylogeny, selection pressure, genotype replacement and molecular clock analyses of dengue viruses imported to Europe by returning travelers
- to compare the 2009 revised WHO dengue case classification to the classical case definition in a non-endemic population (European travelers) in terms of accuracy, sensitivity, simplicity, and acceptability
- to compare clinical manifestations of dengue in a non-endemic population (European travelers) to a endemic population (Sri Lanka) in terms of application of the 2009 revised WHO dengue case classification
- to compare viral genetic factors and clinical manifestations of dengue in a non-endemic population (travelers) and an endemic population (Sri Lanka)
Study affiliation
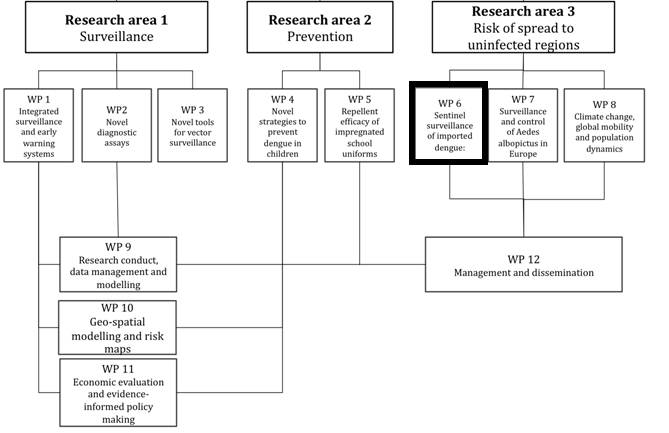
The study will be carried out as part of the DengueTools research initiative funded by the ![]() European commission’s Seventh Framework Programme (FP7). The outline of the initiative’s three main research areas and the associated individual research projects are shown below. This study will be part of “Research area 3: Risk of dengue spreading to uninfected regions” – “Work package (WP) 6″: Sentinel surveillance of imported dengue in returning travelers: trends and virus evolution” (Figure 1).
European commission’s Seventh Framework Programme (FP7). The outline of the initiative’s three main research areas and the associated individual research projects are shown below. This study will be part of “Research area 3: Risk of dengue spreading to uninfected regions” – “Work package (WP) 6″: Sentinel surveillance of imported dengue in returning travelers: trends and virus evolution” (Figure 1).
Study period / time frame
The study will be conducted over a period of two years starting October 2011.
Study population
Eligible study subjects are European residents, who travelled to dengue endemic areas.
Study design
The study will be conducted as a prospective observational multi-centre study.
Inclusion criteria:
Travelers (all age groups) returning from dengue endemic regions who present with an acute febrile illness suspicious of dengue (onset of fever ≤5 days*) within the possible incubation period of a dengue infection (≤21 days).
(* the time frame of ≤5 days after the onset of fever has been assigned as declining viraemia after day 5 is unlikely to enable virus isolation from blood)
Exclusion criteria:
Travelers presenting with concomitant co-infections (e.g. malaria, rickettsial infections, etc.)